Cytoscape simulation of purple membrane biogenesis
The simulation model is based on the current knowledge of the regulatory
and metabolic network responsible for purple membrane biogenesis in
Halobacterium sp. The goal here is to use this model as a prototype
for larger and more complex networks that will result from systems
biology analyses. Furthermore, the simplicity of the simulation model
can be used to introduce high school students to the concept of
Systems Biology. Predictions made by the model can be easily tested
in the laboratory. The attractive aspects of this model pathway lie
in the fact that it is biochemically and genetically tractable and the
phenotypes are colorful and distinct from one perturbation to another.
Purple Membrane Biogenesis in Halobacterium sp.
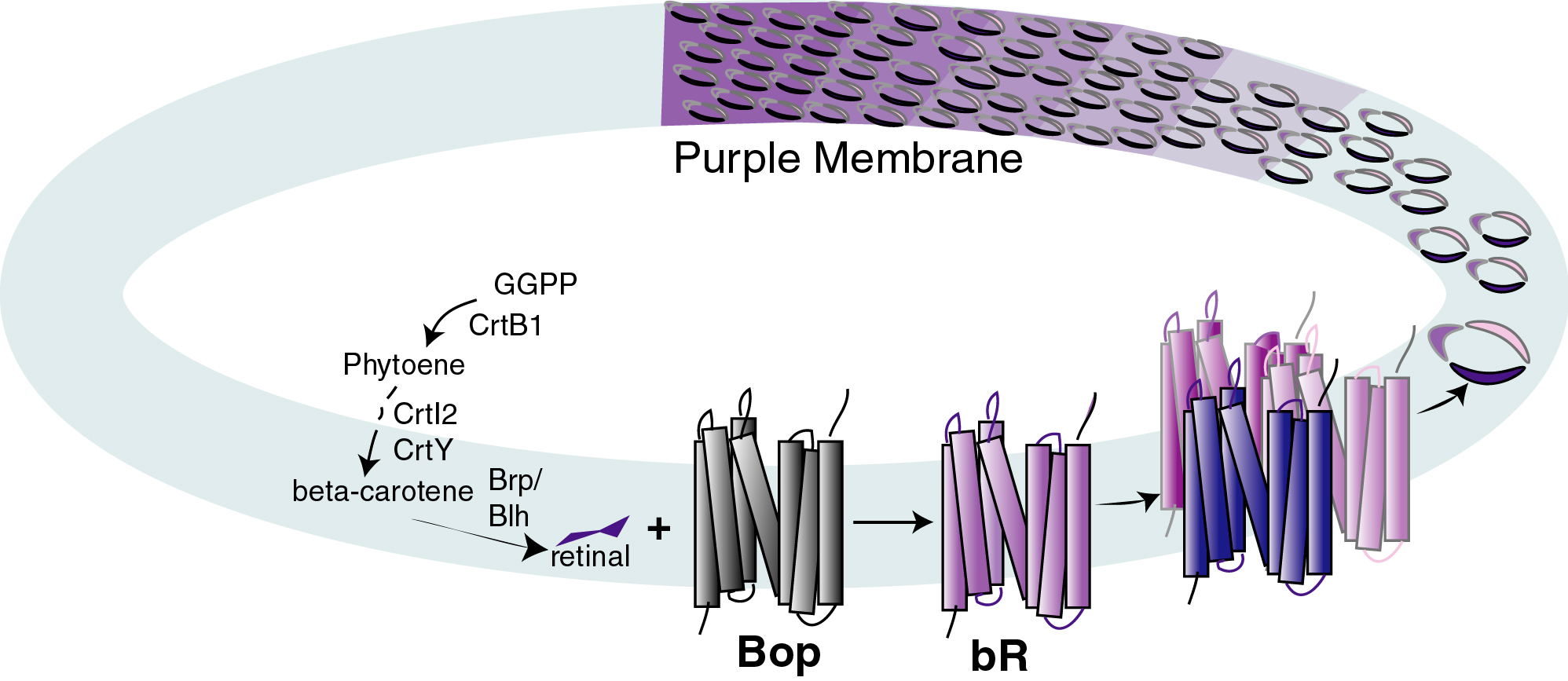
Halobacterium NRC-1, can grow phototrophically via light-driven proton
pumping by bacteriorhodopsin (a 1:1 complex of bacterio-opsin and
retinal) in the purple membrane (Oesterhelt and Stoeckenius, 1973;
Sumper et al., 1976). The proteins for biogenesis of the purple
membrane are coded by the bop gene, specifying bacterio-opsin, and
several nearby genes, e.g. brp, bat, blp, and crtB1, thought to be
involved in synthesis of the chromophore, genetic regulation, or
biogenesis of the membrane (Leong et al., 1988; Ng et al., 2000). The
bat gene encodes a 74-kDa predicted protein (Bat) containing a GAF
(cGMP-binding domain), PAS/PAC (redox-sensing), and DNA-binding HTH
motif. Bat restored oxygen-regulated expression of bop in cis-trans
tests (Yang and DasSarma, 1990; Gropp and Betlach, 1994). Two
frameshift mutations four codons apart within the PAC domain
(233-VVLR-236 to 233-SCCG-236) in Bat were implicated in the
over-expression phenotype of S9 (Baliga et al., 2001). Expression of
bop is induced in mid- to late-exponential phase of growth in response
to both low-oxygen tension and high-light intensity (Yang and
DasSarma, 1990; Shand and Betlach, 1994). Regulation of the bop gene
was dissected further by extensive mutagenesis of the 53-bp minimal
promoter (Gropp et al., 1995; Yang et al., 1996), which identified
three cis-acting elements, a TATA box, for transcription factor and
polymerase recruitment, an RY box, a DNA supercoiling sensitivity
site, and an UAS sequence, the likely site for regulation by Bat.
A genetic analysis of Halobacterium strains NRC-1 (wild type), S9
(overproducer), and SD20 (bat::ISH1 derivative of S9) demonstrated
coordinate regulation of genes, viz. crtB1, brp, and blp, with a
bop-like UAS approximately 40-bp upstream to their respective
transcription start sites (Baliga et al., 2001). Interestingly, the
predicted function of the crtB1 gene product, a phytoene synthase
homolog, is to catalyze the first unique step of retinal and
carotenoid biosynthesis, condensation of two molecules of
geranyl-geranyl pyrophosphate yielding the C-40 isoprenoid compound
phytoene (Armstrong, 1997), while the brp gene product likely
catalyzes the final step in the formation of retinal, oxidative
cleavage of ?-carotene (Peck et al., 2000). Moreover, all
co-regulated genes showed a requirement for a functional bat gene
product suggesting that Bat likely binds the UAS under low-oxygen
tension and high light intensity conditions to coordinate expression
of the structural protein, bacterioopsin, and the chromophore,
retinal. A similar mechanism has been characterized in Arabidopsis
thaliana where phytochrome B (PfrB), a GAF domain containing protein,
gets activated on absorbing red light and translocates into the
nucleus to directly interact with the G-box bound PIF3, a
transcription factor (Martinez-Garcia et al., 2000). The fusion of
the response (GAF and PAS/PAC) and effector (HTH) domains in Bat is
likely due to the lack of requirement of a nuclear translocation step
in Halobacterium sp.
References
- Armstrong, G. A. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu. Rev. Microbiol. 52, 629-659 (1997).
- Baliga, N. S. & DasSarma, S. Saturation mutagenesis of the TATA box and upstream activator sequence in the haloarchaeal bop gene promoter. J. Bacteriol. 181, 2513-2518 (1999).
- Baliga, N. S. & DasSarma, S. Saturation mutagenesis of the haloarchaeal bop gene promoter: identification of DNA supercoiling sensitivity sites and absence of TFB recognition element and UAS enhancer activity. Mol. Microbiol. 36, 1175-1183 (2000).
- Gropp, F. & Betlach, M. C. The bat gene of Halobacterium halobium encodes a trans-acting oxygen inducibility factor. Proc. Natl Acad. Sci. U.S.A. 91, 5475-5479 (1994).
- Gropp, F., Gropp, R. & Betlach, M. C. Effects of upstream deletions on light- and oxygen-regulated bacterio-opsin gene expression in Halobacterium halobium. Mol. Microbiol. 16, 357-364 (1995).
- Martinez-Garcia, J. F., Huq, E. & Quail, P. H. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859-863 (2000).
- Ng, W. V. et al. From the cover: genome sequence of Halobacterium species NRC-1. Proc. Natl Acad. Sci. U.S.A. 97, 12176-12181 (2000).
- Oesterhelt, D. & Stoeckenius, W. Functions of a new photoreceptor membrane. Proc. Natl Acad. Sci. U.S.A. 70, 2853-2857 (1973).
- Peck, R. F., Echavarri-Erasun, C., Johnson, E. A., Ng, W. V., Kennedy, S. P., Hood, L., DasSarma, S. & Krebs, M. P. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J. Biol. Chem. epub ahead of print (2000).
- Shand, F. R. & Betlach, M. C. Expression of the bop gene cluster of Halobacterium halobium is induced by low oxygen tension and by light. J. Bacteriol. 173, 4692-4699 (1991).
- Sumper, M., Reitmeier, H. & Oesterhelt, D. Biosynthesis of the purple membrane of halobacteria. Angew. Chem. Int. Ed. Engl. 16, 187-194 (1976).
- Yang, C.-F. & DasSarma, S. Transcriptional induction of purple membrane and gas vesicle synthesis in the archaebacterium Halobacterium halobium is blocked by a DNA gyrase inhibitor. J. Bacteriol. 172, 4118-4121 (1990).
- Yang, C.-F., Kim, J.-M., Molinari, E. & DasSarma, S. Genetic and topological analyses of the bop promoter of Halobacterium halobium: stimulation by DNA supercoiling and non-B-DNA structure. J. Bacteriol. 178, 840-845 (1996).

